* below = equal contributions.
I’ve worked on a diverse set of topics, but underlying all my research is a wish for a predictive understanding of biomolecular structure ensembles. My work has leveraged novel tools such as high-throughput experiments, deep learning, and crowdsourcing, while remaining grounded in biochemical principles and prioritizing testable predictions.
Predicting multiple conformations of known and putative fold-switching proteins
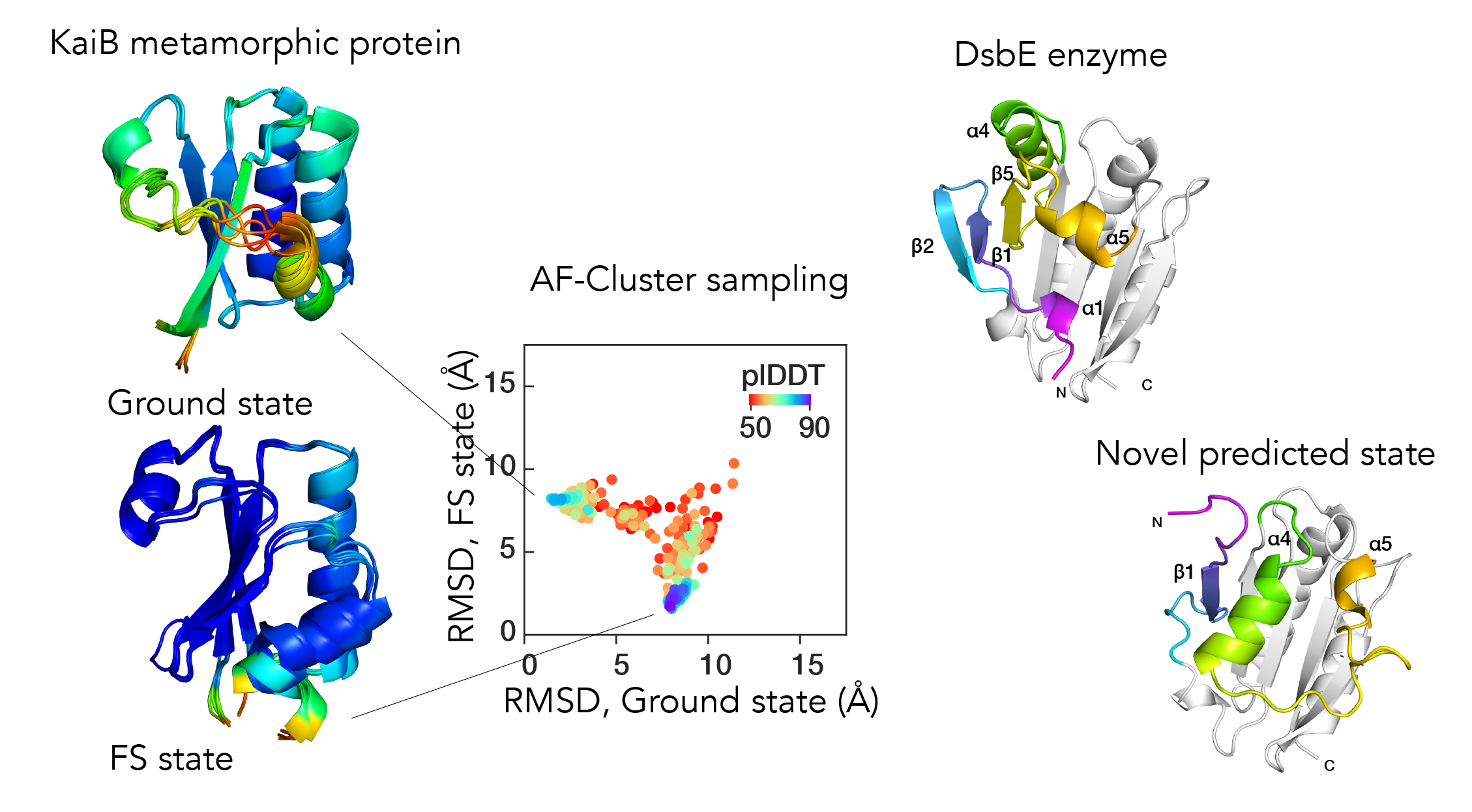 AlphaFold2 (AF2) has revolutionized structural biology by accurately predicting single structures of proteins, but biological function is rooted in a protein’s ability to sample different conformational substates. We demonstrated that simply clustering an input multiple sequence alignment (MSA) by sequence similarity enables AF2 to sample alternate states of known metamorphic proteins and score these states with high confidence. We used information gained from the clustering to design and validate a minimal set of point mutations to flip the population of KaiB from one state to the other.
We used our clustering method, AF-cluster, to screen for alternate states in protein families without known fold-switching, and identified a putative alternate state for the oxidoreductase Mpt53, secreted by *M. tuberculosis* during infection. Similarly to KaiB, Mpt53 is predicted to switch between a thioredoxin-like fold and a novel fold.
AlphaFold2 (AF2) has revolutionized structural biology by accurately predicting single structures of proteins, but biological function is rooted in a protein’s ability to sample different conformational substates. We demonstrated that simply clustering an input multiple sequence alignment (MSA) by sequence similarity enables AF2 to sample alternate states of known metamorphic proteins and score these states with high confidence. We used information gained from the clustering to design and validate a minimal set of point mutations to flip the population of KaiB from one state to the other.
We used our clustering method, AF-cluster, to screen for alternate states in protein families without known fold-switching, and identified a putative alternate state for the oxidoreductase Mpt53, secreted by *M. tuberculosis* during infection. Similarly to KaiB, Mpt53 is predicted to switch between a thioredoxin-like fold and a novel fold.
I’m really excited about what AF-cluster and future methodological improvements could enable – the ability to systematically screen for interesting novel mechanisms based on multiple conformational substates, and discover new biology from the ground up.
Wayment-Steele, H.K.*, Ojoawo, A.*, … Kern, D. Predicting multiple conformations via sequence clustering and AlphaFold2. Nature (2023). doi
Designing stabilized mRNA therapeutics
 One of the greatest global health challenges that faces biomedical engineering is the development of refrigerator-stable vaccines to enable more equitable distribution. Vaccines based on messenger RNA (mRNA) have immense promise, yet persistent concerns regarding their thermostability. A largely unexplored strategy to reduce mRNA hydrolysis is to design mRNAs that form double-stranded regions, which are protected from in-line cleavage and enzymatic degradation, while coding for the same proteins. The amount of stabilization that this strategy can deliver and the most effective algorithmic approach to achieve stabilization were poorly understood. My computational work, in collaboration with a team from the Barna lab at Stanford and the Eterna RNA design project, led to mRNA designs that when synthesized and experimentally characterized, had a 2.5-fold increase in half-life over conventional methods, and, to our surprise, increased protein expression over conventional designs. We used these data to launch a crowdsourced machine-learning challenge on the platform Kaggle, in which over 1600 teams collaborated to develop highly accurate models for predicting RNA degradation (Wayment-Steele, 2021, arXiv). We anticipate this work will be formative for guiding future therapeutic and vaccine development in potency and stability.
One of the greatest global health challenges that faces biomedical engineering is the development of refrigerator-stable vaccines to enable more equitable distribution. Vaccines based on messenger RNA (mRNA) have immense promise, yet persistent concerns regarding their thermostability. A largely unexplored strategy to reduce mRNA hydrolysis is to design mRNAs that form double-stranded regions, which are protected from in-line cleavage and enzymatic degradation, while coding for the same proteins. The amount of stabilization that this strategy can deliver and the most effective algorithmic approach to achieve stabilization were poorly understood. My computational work, in collaboration with a team from the Barna lab at Stanford and the Eterna RNA design project, led to mRNA designs that when synthesized and experimentally characterized, had a 2.5-fold increase in half-life over conventional methods, and, to our surprise, increased protein expression over conventional designs. We used these data to launch a crowdsourced machine-learning challenge on the platform Kaggle, in which over 1600 teams collaborated to develop highly accurate models for predicting RNA degradation (Wayment-Steele, 2021, arXiv). We anticipate this work will be formative for guiding future therapeutic and vaccine development in potency and stability.
We had a blast talking with 50 Years about this work on their podcast.
I was also delighted to share this work with Washington High in Fremont, CA at their TedX conference.
Wayment-Steele, H. K., Kim, D. S., Choe, C. A., Nicol, J. J., Wellington-Oguri, R., Watkins, A. M., . . . Das, R. (2021). Theoretical basis for stabilizing messenger RNA through secondary structure design. Nucleic Acids Res, 49(18), 10604-10617.
Leppek, K.*, Byeon, G. W.*, Kladwang, W.*, Wayment-Steele, H. K.*, Kerr, C. H.*, Xu, A. F., . . . Dormitzer, P., Solorzano, A., Barna, M., Das, R. (2021). Combinatorial optimization of mRNA structure, stability, and translation for RNA-based therapeutics. Nature Communications.
Wayment-Steele, H. K.*, Kladwang, W.*, Watkins, A. M.*, Kim, D. S.*, Tunguz, B.*, Reade, W., … & Das, R. (2021). Predictive models of RNA degradation through dual crowdsourcing. Nature Machine Intelligence (In Press).
Improving RNA ensemble prediction and design
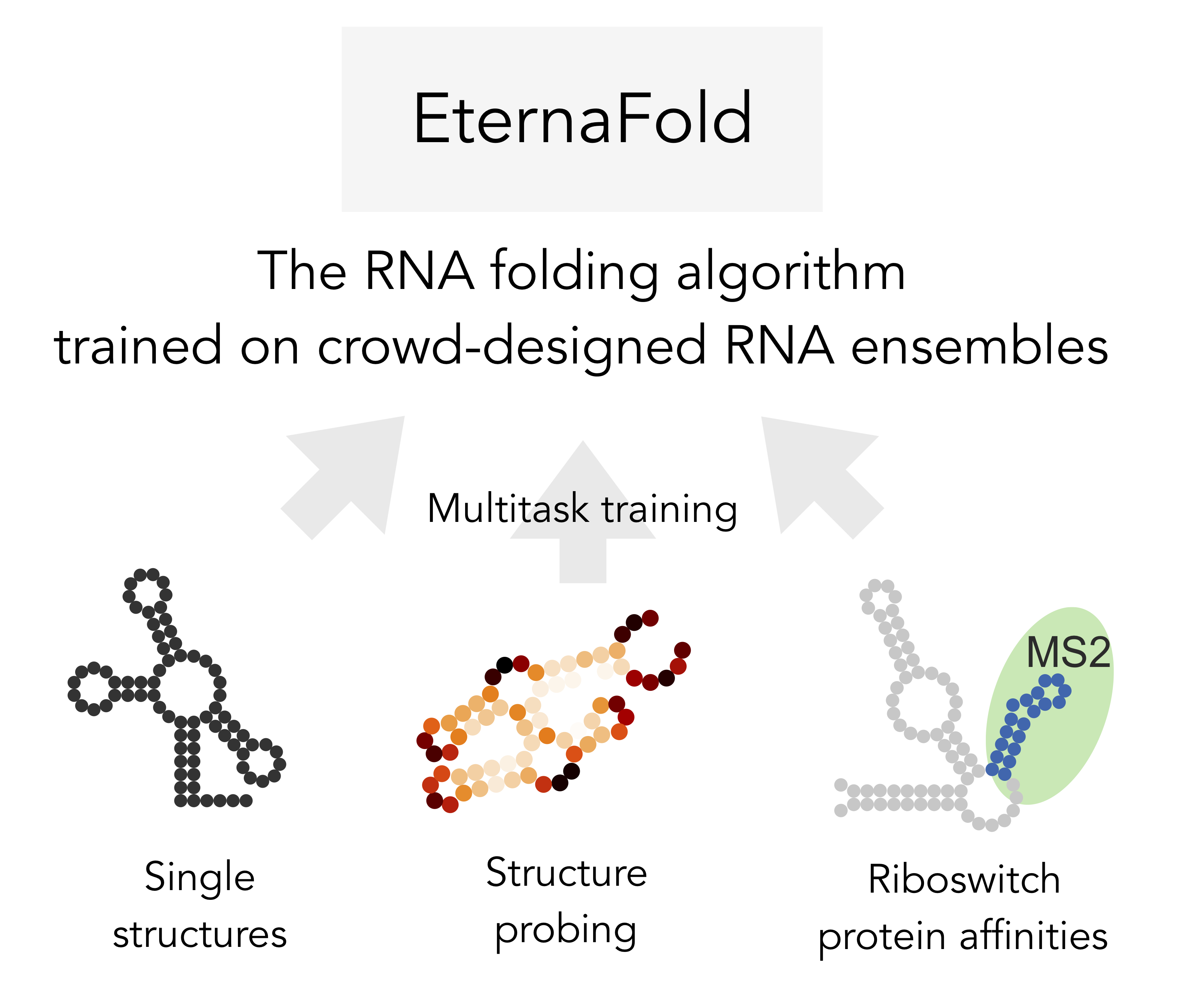 The computer-aided study and design of RNA molecules is increasingly important across a range of disciplines, yet little was known about the accuracy of commonly used structure prediction packages in real-world tasks. For example, riboswitches are RNA elements that recognize diverse chemical and biomolecular inputs and transduce this recognition process to genetic, fluorescent, and other engineered outputs using RNA conformational changes. I developed the first high-throughput, independent evaluation of all commonly-used structure prediction algorithms using two separate thermodynamic prediction tasks: 1) predicting unpaired probabilities of individual nucleotides through chemical mapping data, and 2) predicting riboswitch activity. I further developed a multitask-learning-based model trained from these data, EternaFold, which demonstrated improved performance that generalized to diverse external datasets, including complete viral genomes probed in vivo and synthetic designs modeling mRNA vaccines.
The computer-aided study and design of RNA molecules is increasingly important across a range of disciplines, yet little was known about the accuracy of commonly used structure prediction packages in real-world tasks. For example, riboswitches are RNA elements that recognize diverse chemical and biomolecular inputs and transduce this recognition process to genetic, fluorescent, and other engineered outputs using RNA conformational changes. I developed the first high-throughput, independent evaluation of all commonly-used structure prediction algorithms using two separate thermodynamic prediction tasks: 1) predicting unpaired probabilities of individual nucleotides through chemical mapping data, and 2) predicting riboswitch activity. I further developed a multitask-learning-based model trained from these data, EternaFold, which demonstrated improved performance that generalized to diverse external datasets, including complete viral genomes probed in vivo and synthetic designs modeling mRNA vaccines.
Give EternaFold a spin!
Wayment-Steele, H.K., Kladwang, W., Strom, A.I., Lee, J., Treuille, A., Eterna Participants, Das, R. (2022) RNA secondary structure packages evaluated and improved by high-throughput experiments. Nature Methods (In Press).
Wayment-Steele, H., Wu, M., Gotrik, M., & Das, R. (2019). Evaluating riboswitch optimality. Methods Enzymol, 623, 417-450. doi:10.1016/bs.mie.2019.05.028
Andreasson, J. O., Gotrik, M. R., Wu, M. J., Wayment-Steele, H. K., Kladwang, W., Portela, F., … & Greenleaf, W. J. (2022). Crowdsourced RNA design discovers diverse, reversible, efficient, self-contained molecular sensors. Proc. Natl. Acad. Sci. (In Press).
Unsupervised deep learning for improved protein folding kinetic models and protein-protein interactions
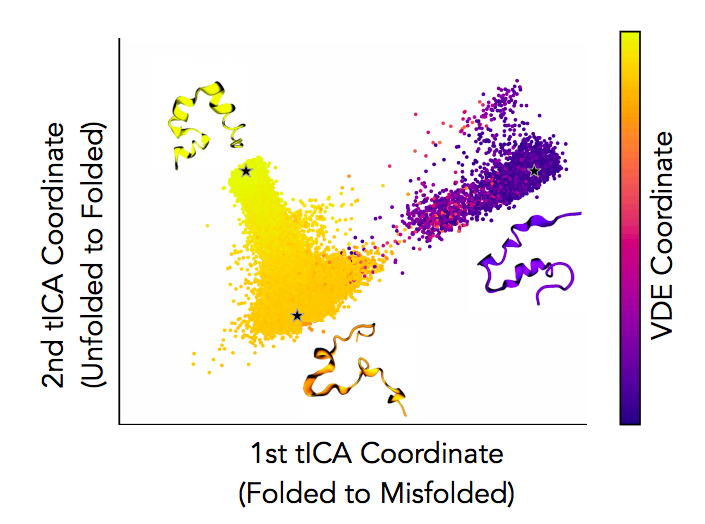 Molecular dynamics (MD) simulations offer the potential to understand atomistic details of protein dynamics. As MD simulations are able to sample increasing length- and time-scales, new theoretical and statistical methods are needed to parse the resulting data to extract meaningful states and kinetic rate constants. We leveraged a classic framework from unsupervised learning, the variational autoencoder, to develop an improved approach for extracting long-timescale processes from MD data. I applied these approaches to model protein-protein interaction dissociation timescales and geometries of a junctured-DNA tweezers as a generic platform to enable real-time observation, at the single- molecule level, of biomolecular interactions.
Molecular dynamics (MD) simulations offer the potential to understand atomistic details of protein dynamics. As MD simulations are able to sample increasing length- and time-scales, new theoretical and statistical methods are needed to parse the resulting data to extract meaningful states and kinetic rate constants. We leveraged a classic framework from unsupervised learning, the variational autoencoder, to develop an improved approach for extracting long-timescale processes from MD data. I applied these approaches to model protein-protein interaction dissociation timescales and geometries of a junctured-DNA tweezers as a generic platform to enable real-time observation, at the single- molecule level, of biomolecular interactions.
Kostrz, D., Wayment-Steele, H. K., Wang, J. L., Follenfant, M., Pande, V. S., Strick, T. R., & Gosse, C. (2019). A modular DNA scaffold to study protein-protein interactions at single-molecule resolution. Nat Nanotechnol, 14(10), 988-993.
HK Wayment-Steele & VS Pande (2018). Note: Variational Encoding of Protein Dynamics Benefits from Maximizing Latent Autocorrelation. J. Chem. Phys. 149, 216101.
MM Sultan, HK Wayment-Steele, & VS Pande. Transferable Neural Networks for Enhanced Sampling of Protein Dynamics. J. Chem. Theor. Comp. 2018 14 (4), 1887-1894.
CX Hernández*, HK Wayment-Steele*, MM Sultan*, BE Husic, & VS Pande. (2017). Variational Encoding of Complex Dynamics. Phys. Rev. E. 97, 062412. *Equal contributions
Studying self-assembly in DNA origami
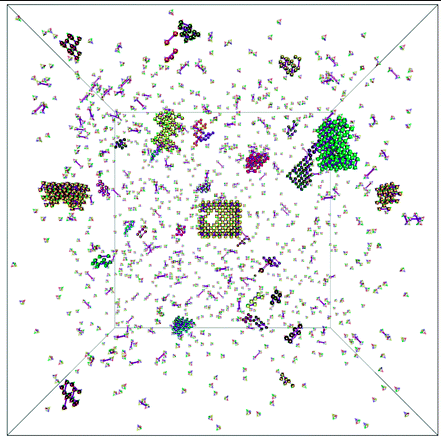 DNA origami nanomaterials hold promise in fields ranging from drug discovery to nanoelectronics for their durability and tunability, yet their yield can be low due to off-pathway assembly. An understanding of the thermodynamics of the assembly process would allow us to improve the yield and design more complex structures. In this work, I studied one design principle for a "DNA brick" system, using larger "boundary bricks" at the edges of structures, and proposed a mechanism of how this design leads to more robust assembly using Monte Carlo simulation.
DNA origami nanomaterials hold promise in fields ranging from drug discovery to nanoelectronics for their durability and tunability, yet their yield can be low due to off-pathway assembly. An understanding of the thermodynamics of the assembly process would allow us to improve the yield and design more complex structures. In this work, I studied one design principle for a "DNA brick" system, using larger "boundary bricks" at the edges of structures, and proposed a mechanism of how this design leads to more robust assembly using Monte Carlo simulation.
Funded by the Churchill Scholarship.
HK Wayment-Steele, D Frenkel, A Reinhardt. Investigating the role of boundary bricks in DNA brick self-assembly. Soft Matter 2017 13 (8), 1670-1680.
Understanding the neurotoxic effects of aluminum ions on cell membranes
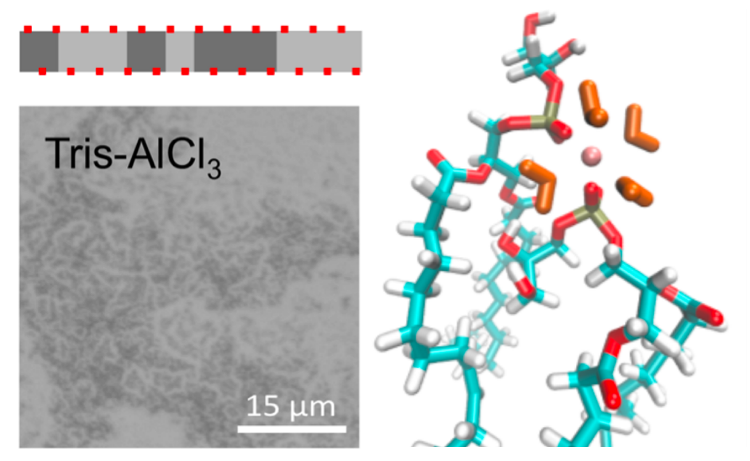 Dissolved aluminum (Al3+) is abundant in our surroundings yet is known to be neurotoxic in acute doses, and chronic exposure is increasingly correlated with Alzheimers-like symptoms. In this work, I demonstrated that Al3+ ions cause cell membranes to adopt a gel-like structure and reduce their diffusivity, which is highly detrimental to essential membrane functions. I contributed fluorescence microscopy, surface gravimetric measurements (QCM-D), and molecular dynamics simulations in this work.
Dissolved aluminum (Al3+) is abundant in our surroundings yet is known to be neurotoxic in acute doses, and chronic exposure is increasingly correlated with Alzheimers-like symptoms. In this work, I demonstrated that Al3+ ions cause cell membranes to adopt a gel-like structure and reduce their diffusivity, which is highly detrimental to essential membrane functions. I contributed fluorescence microscopy, surface gravimetric measurements (QCM-D), and molecular dynamics simulations in this work.
Funded by the Goldwater Scholarship & Beckman Scholarship.
Completed as a visiting scholar at Chalmers University of Technology (Gothenburg, Sweden), as a guest of S. Svedhem, and as a visiting scholar at the University of Göttingen (Göttingen, Germany), as a guest of A. Kunze.
Wayment-Steele, H. K., Jing, Y., Swann, M. J., Johnson, L. E., Agnarsson, B., Svedhem, S., & Kunze, A. (2016). Effects of Al3+ on Phosphocholine and Phosphoglycerol Containing Solid Supported Lipid Bilayers. Langmuir, 32(7), 1771-1781.
Agnarsson, B., Wayment-Steele, H. K., Höök, F., & Kunze, A. (2016). Monitoring of single and double lipid membrane formation with high spatiotemporal resolution using evanescent light scattering microscopy. Nanoscale, 8(46), 19219-19223.
Characterizing surface interactions in materials for alternative energy
Wayment-Steele, H. K., Johnson, L. E., Tian, F., Dixon, M. C., Benz, L., & Johal, M. S. (2014). Monitoring N3 dye adsorption and desorption on TiO2 surfaces: a combined QCM-D and XPS study. ACS applied materials & interfaces, 6(12), 9093-9099.
Tian, F., Cerro, A. M., Mosier, A. M., Wayment-Steele, H. K., … & Benz, L. (2014). Surface and stability characterization of a nanoporous ZIF-8 thin film. J. Phys. Chem. C, 118(26), 14449-14456.
